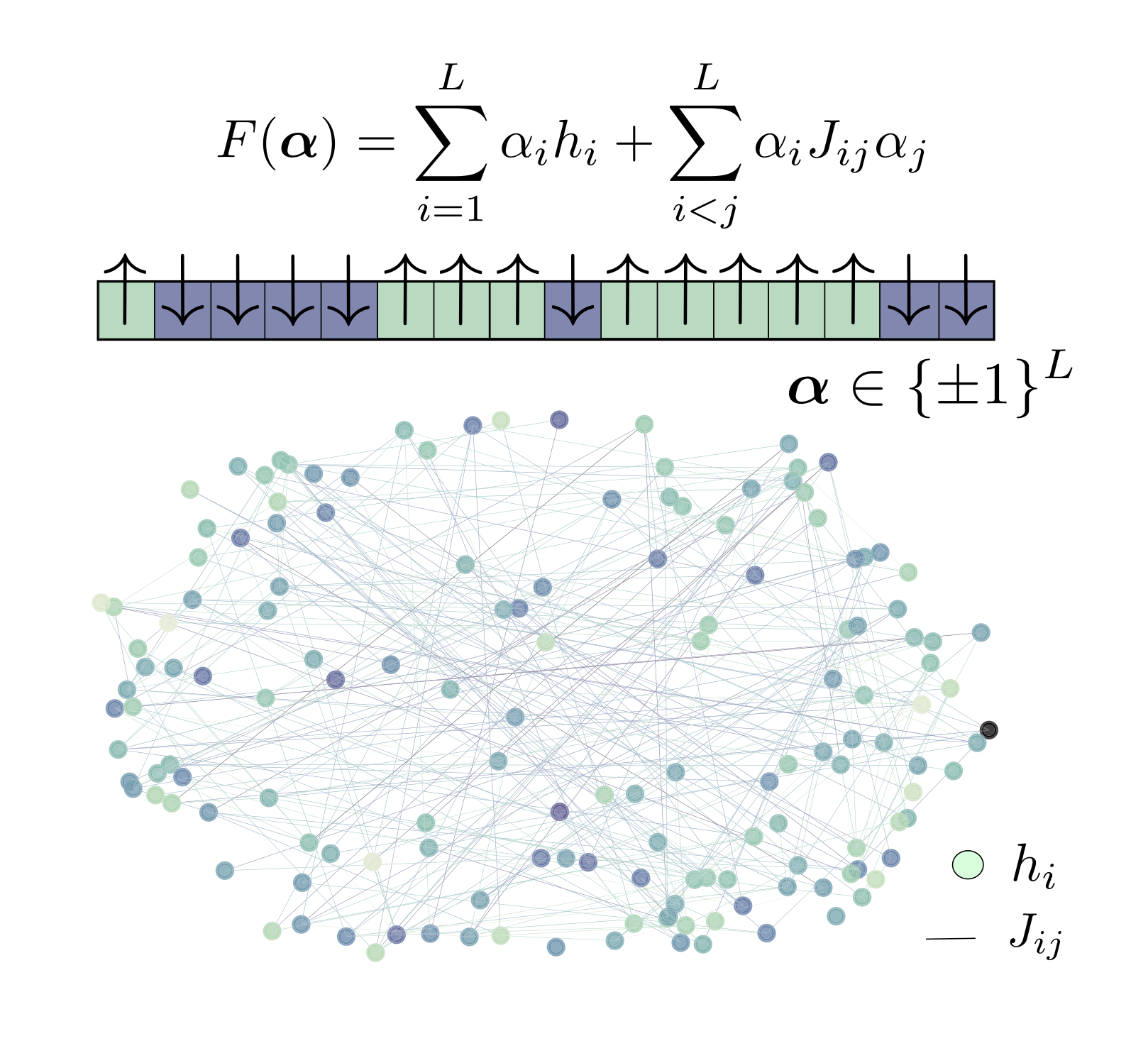
I am an assistant professor in the Machine Learning Department and the Department of Mathematical Sciences at CMU. I lead a focused team that studies the algorithmic foundations of generative models and their application to problems across science and engineering. Previously, I was a Courant Instructor at the Courant Institute of Mathematical Sciences working with Eric Vanden-Eijnden. I completed my PhD in applied mathematics at Harvard co-advised by Jean-Jacques Slotine and Chris Rycroft. A longer bio is here.
My work has been supported by a Department of Energy Computational Science Graduate Fellowship, a Fulbright Research Fellowship, a Google Student Researcher Award, and an NSF Mathematical Sciences Postdoctoral Research Fellowship.
Research /
Publications /
Code /
Teaching /
Application Materials
Research Group
The goal of our group is to develop a mathematical theory of generative artificial intelligence and to demonstrate that the resulting techniques can solve scientific problems previously thought intractable. We design simple and elegant algorithmic frameworks that elucidate why existing approaches work, reveal principled new methods, and scale to achieve state-of-the-art performance in practice.
Questions we are currently studying include:
Foundations of generative modeling: Why do flow and diffusion models work so well, and how do we move beyond the current paradigm?
In this direction, we have developed:
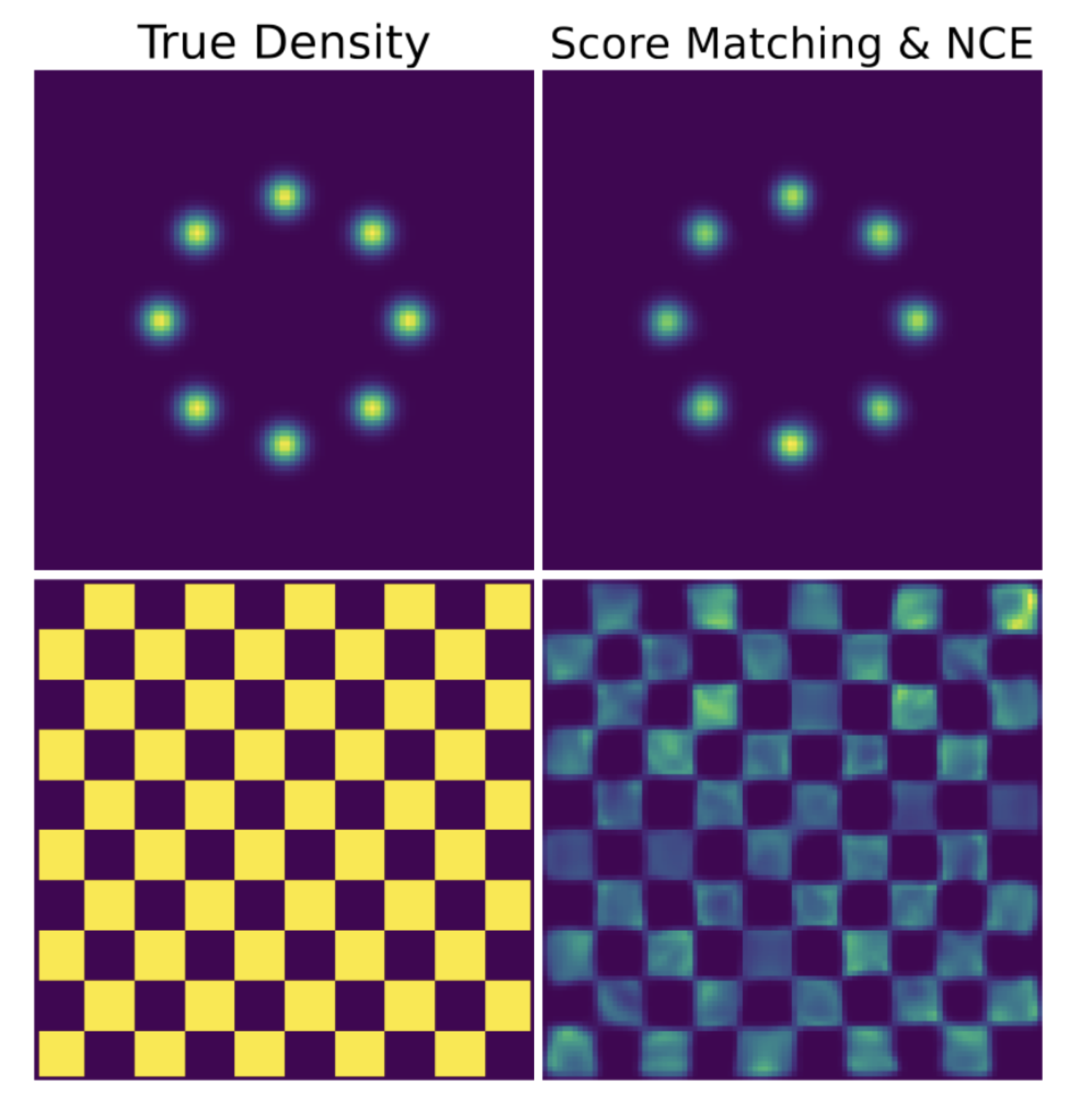
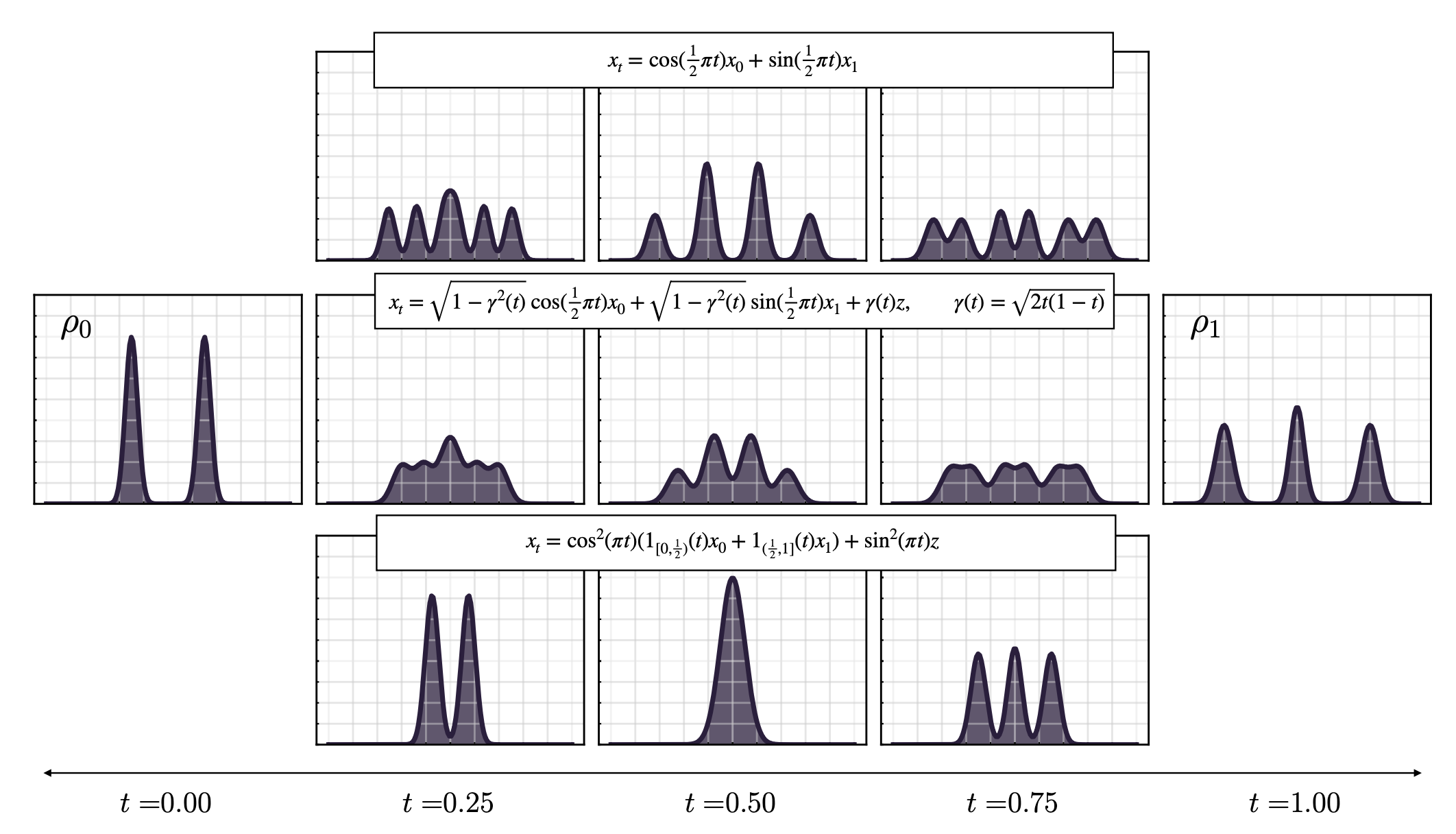
- Stochastic interpolants, a mathematical framework that unifies and underlies modern flow and diffusion models
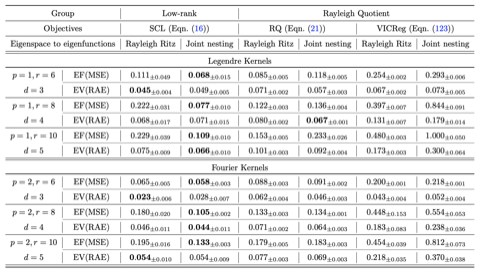
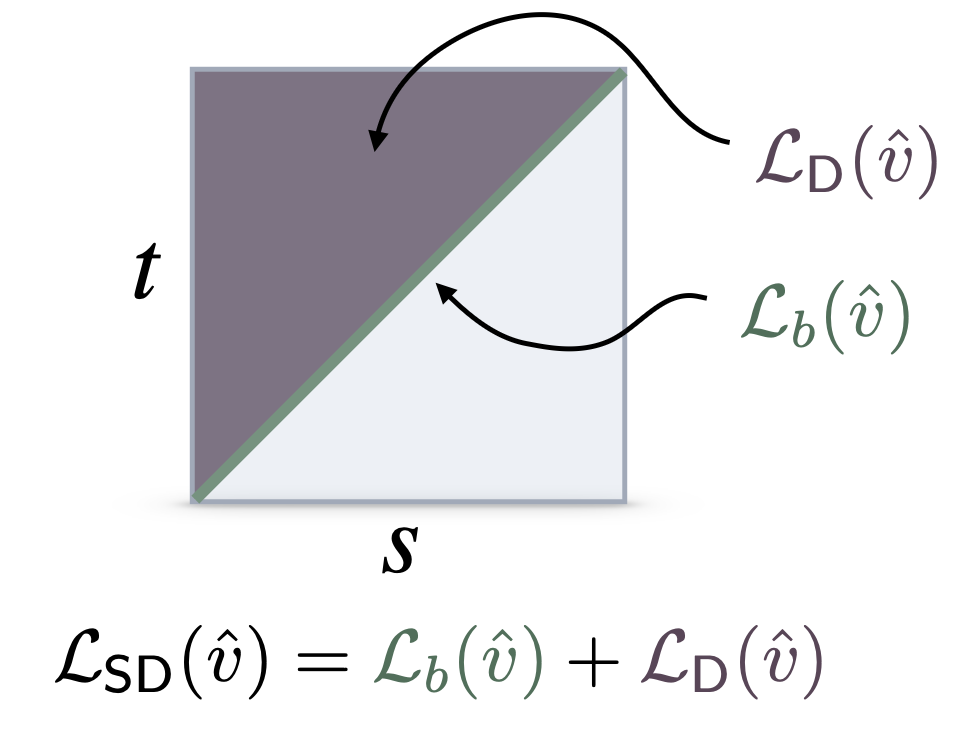
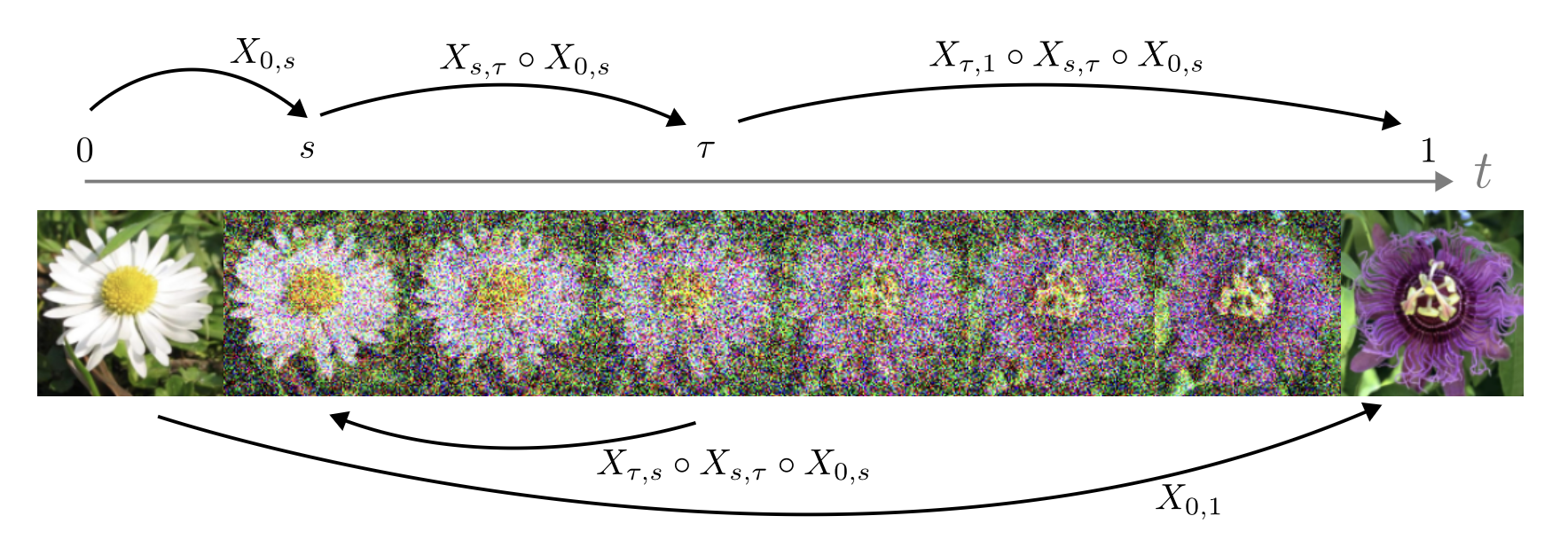
- Flow maps, a theory of accelerated generative models that explains and extends consistency models
Current topics of interest include controlled generation, fine-tuning, inference-time guidance, accelerated sampling, and discrete generation.
Generative models for science and engineering: How can we solve scientific problems that are fundamentally impossible with traditional computational techniques?
We pursue applications in physics, chemistry, biology, vision, and robotics. In each case, we collaborate closely with domain experts to ensure we are solving real problems, as well as targeting challenges that could not be addressed with known domain-specific tools.
In this direction, we have developed:
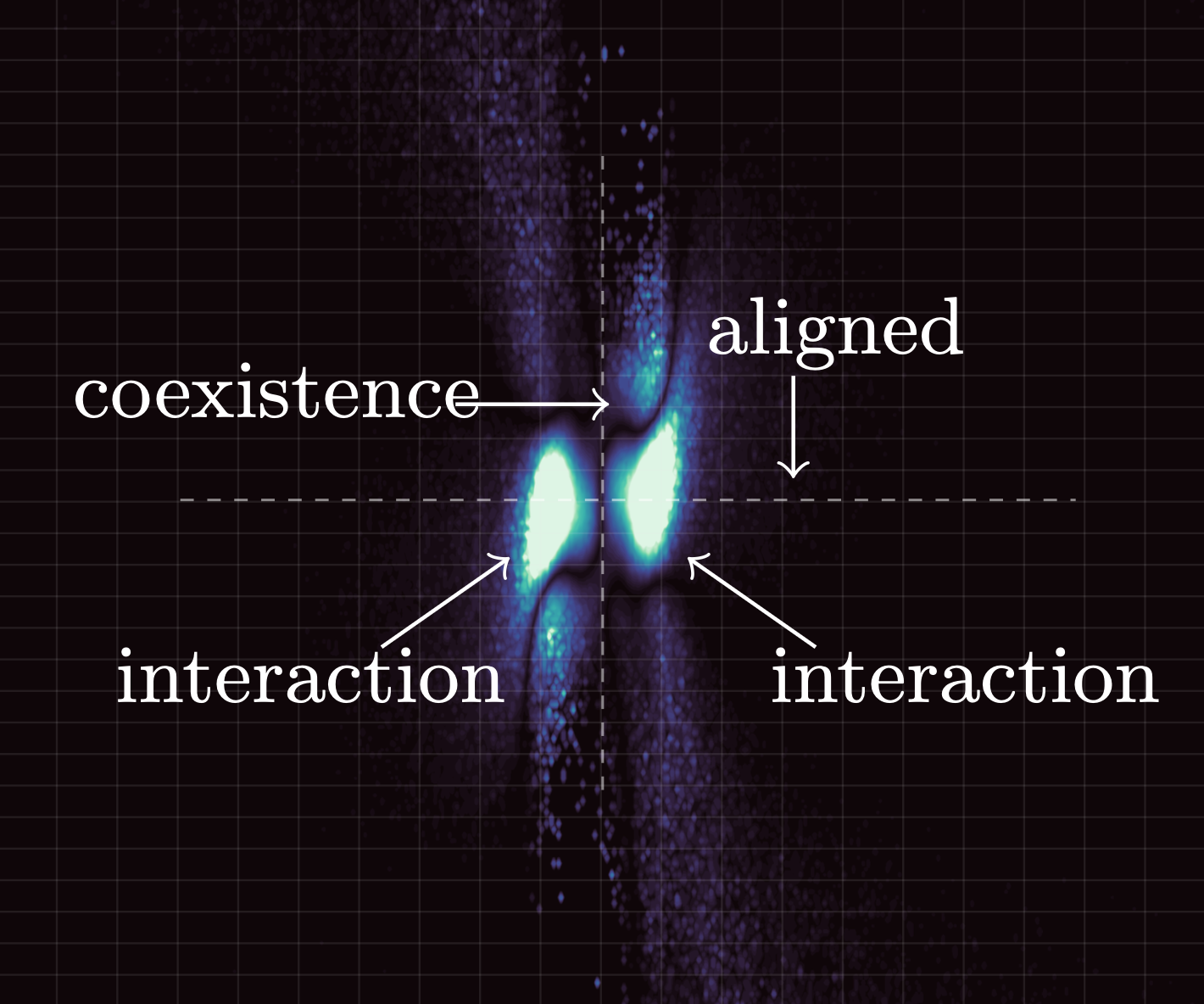
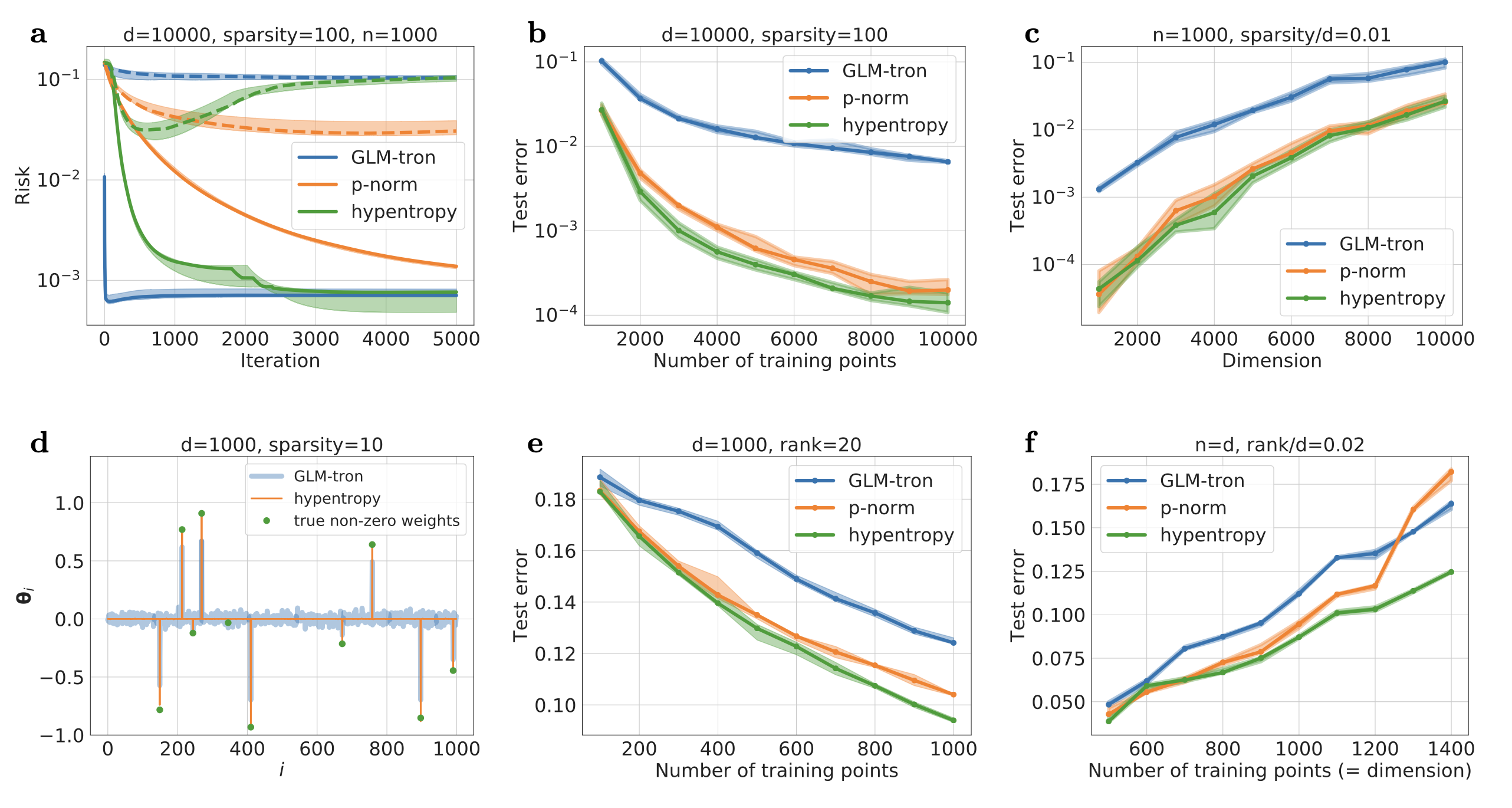
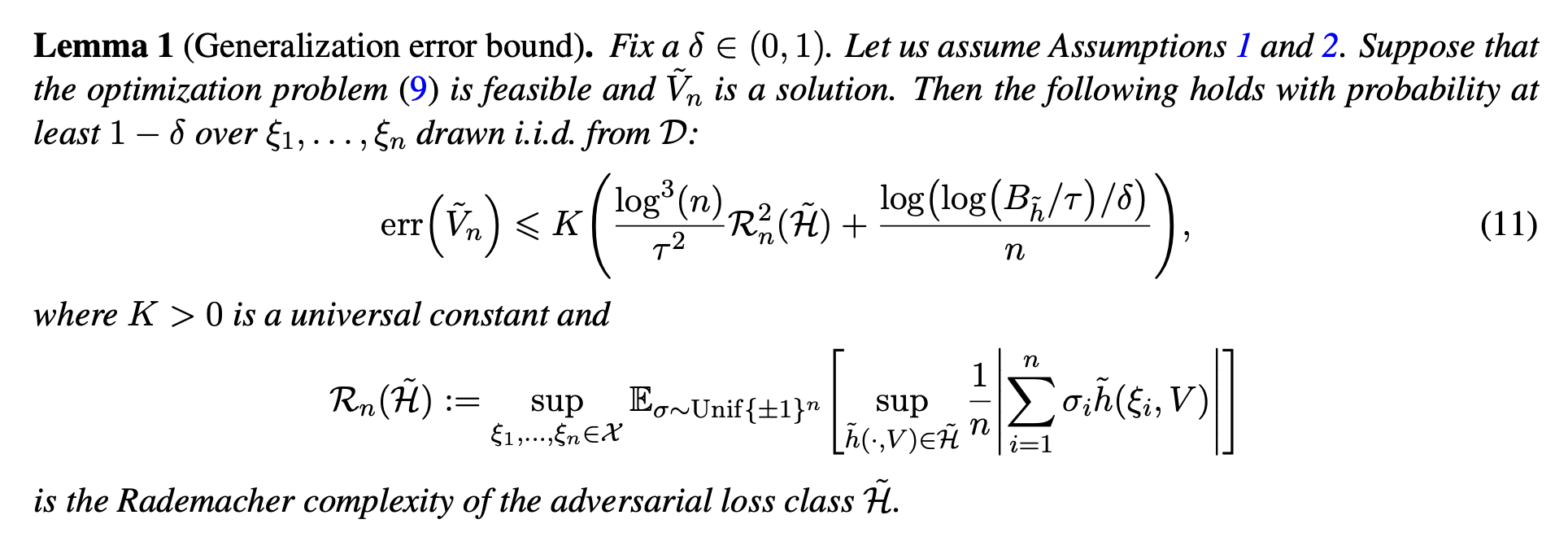
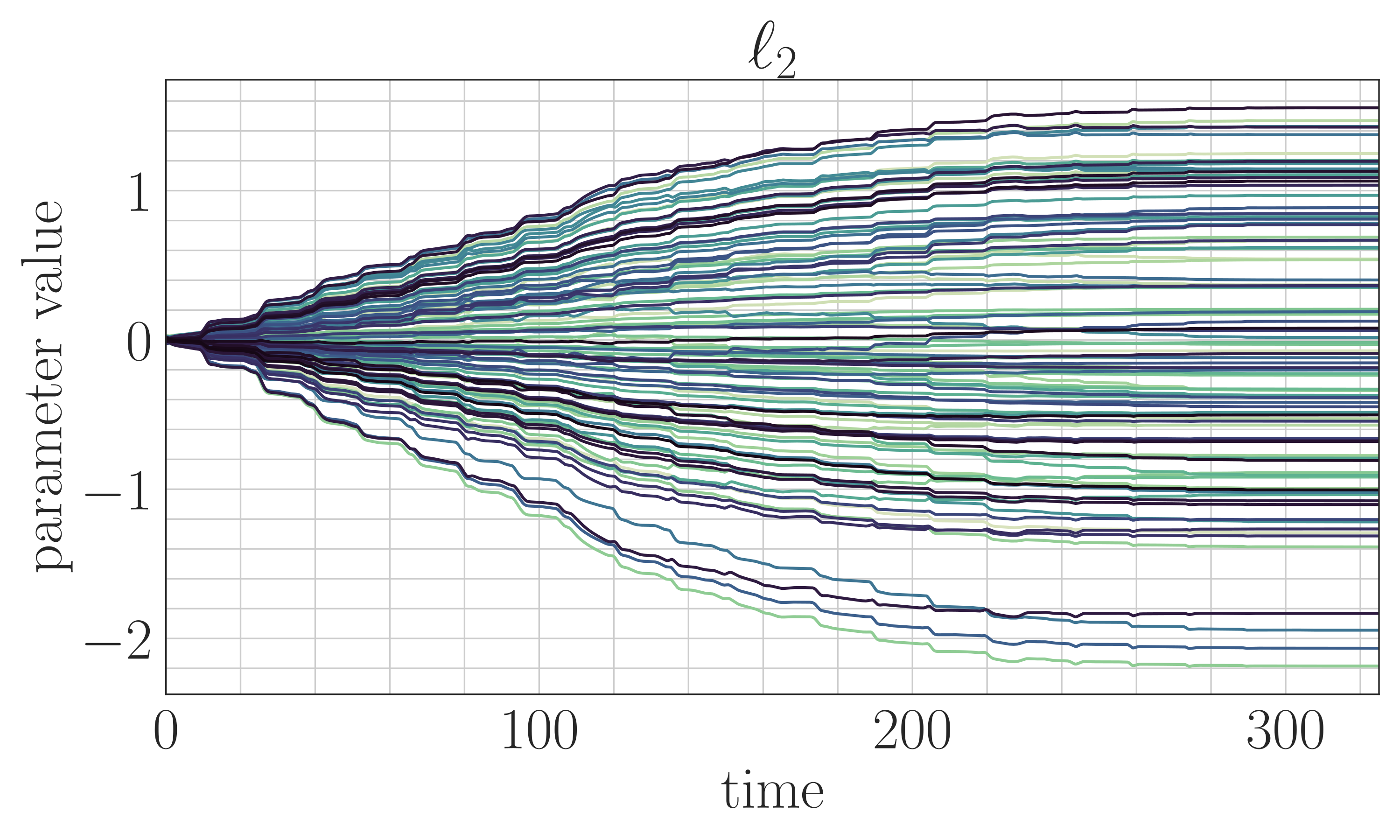
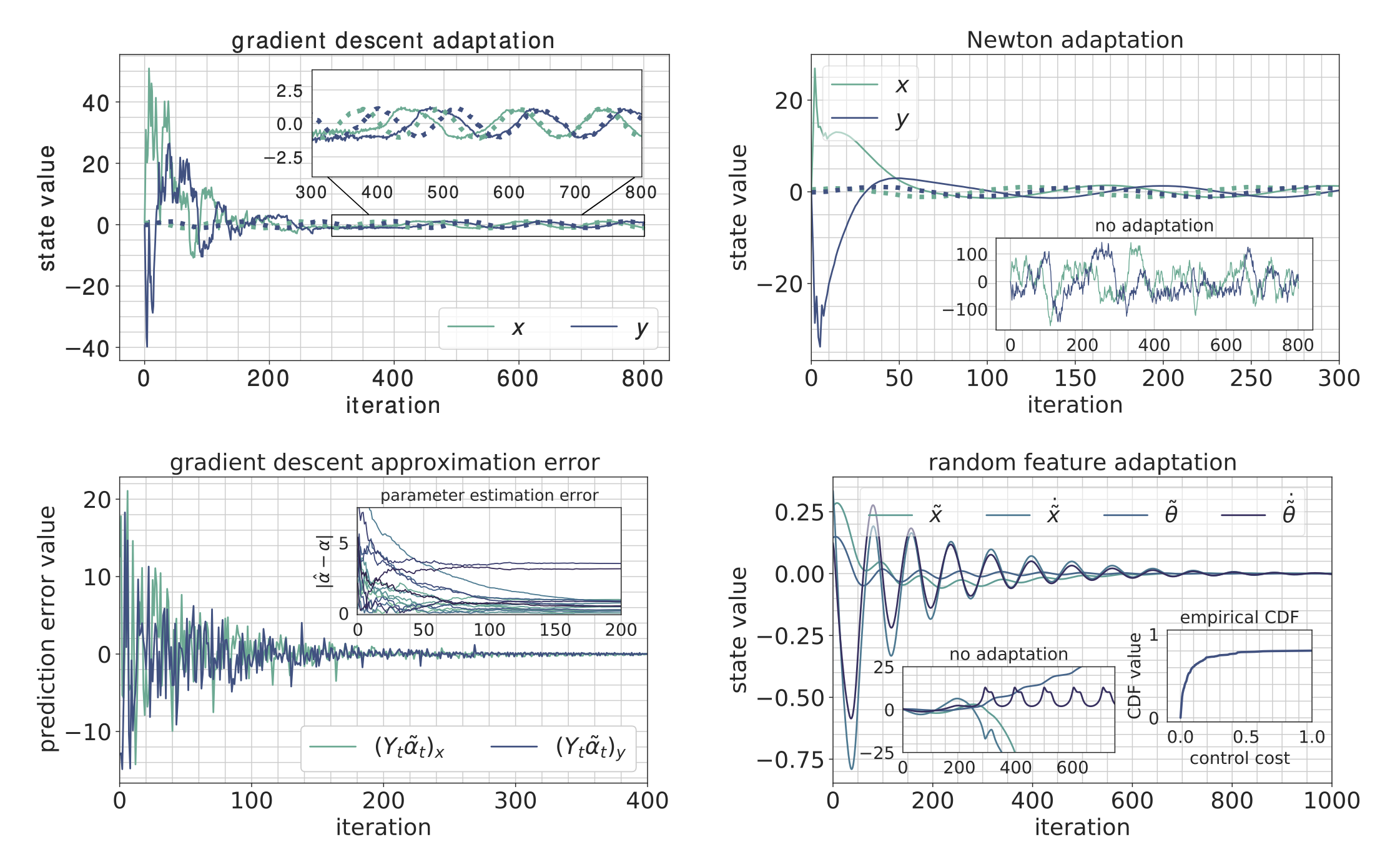
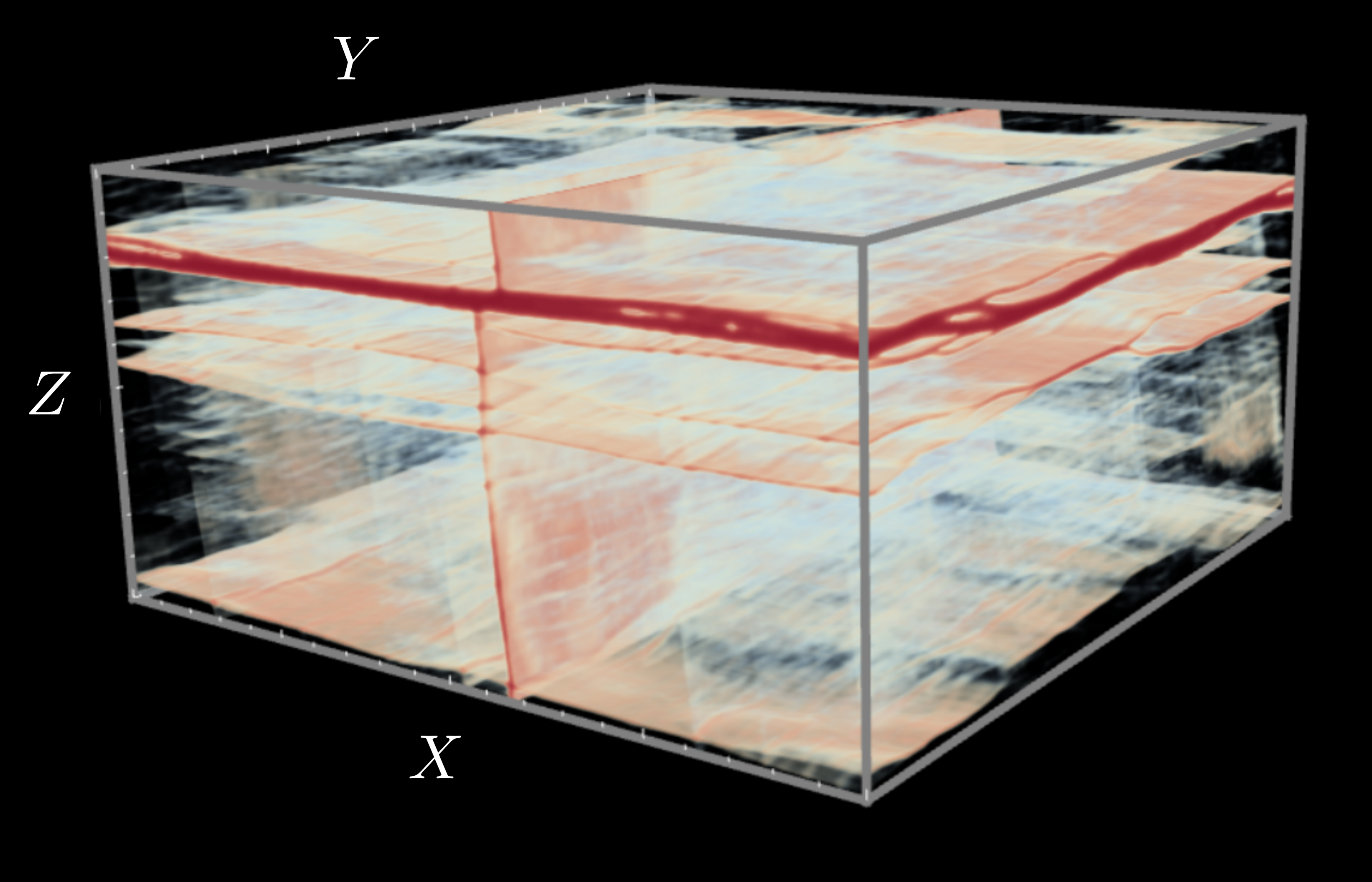
- Methods for entropy production estimation in non-equilibrium systems that scale to tens of thousands of dimensions
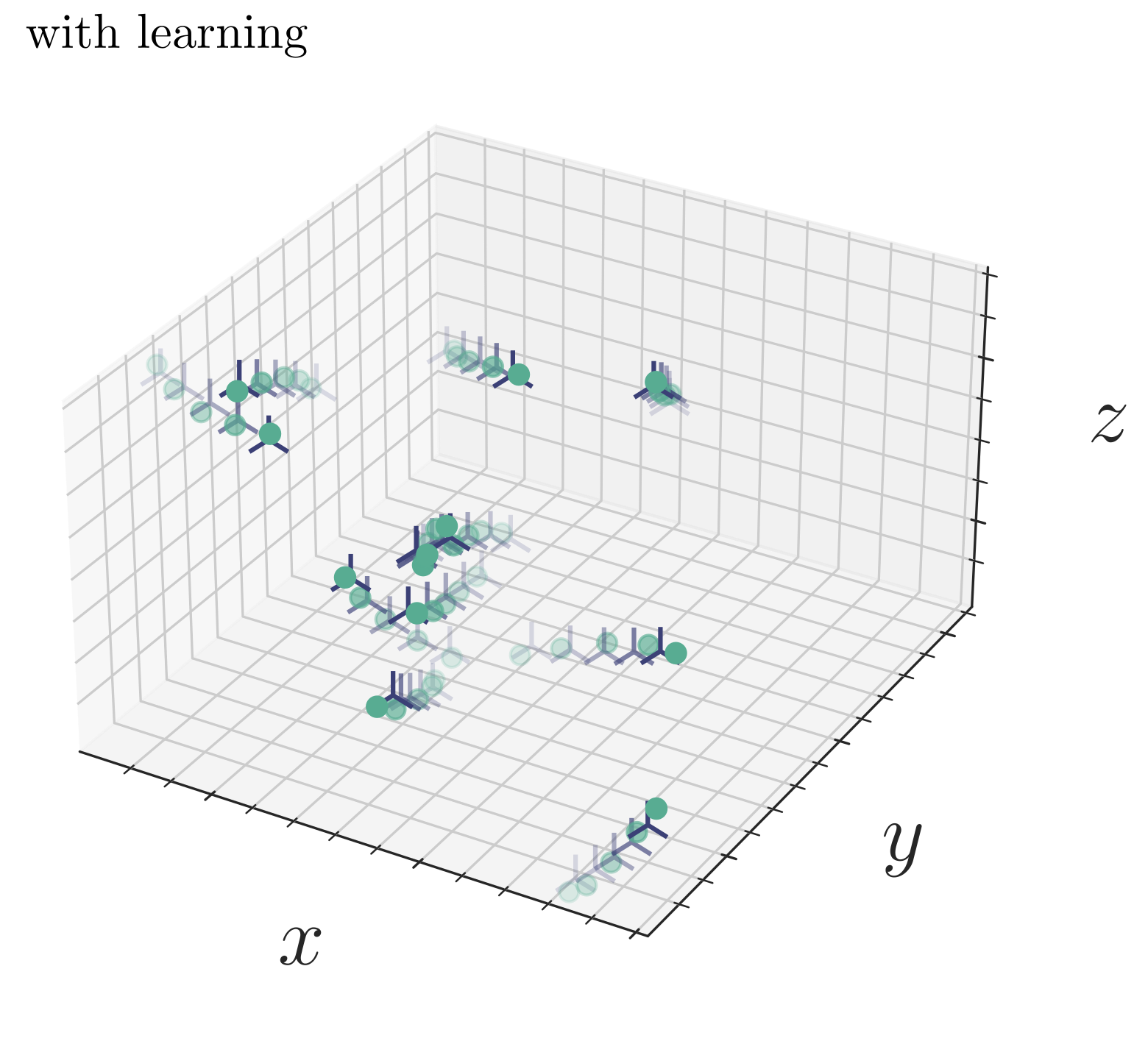
- A critical analysis of generative policies in robotic control, showing they work for different reasons than commonly understood
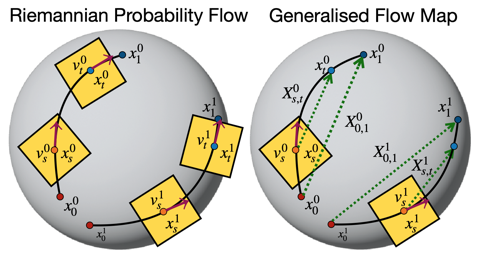
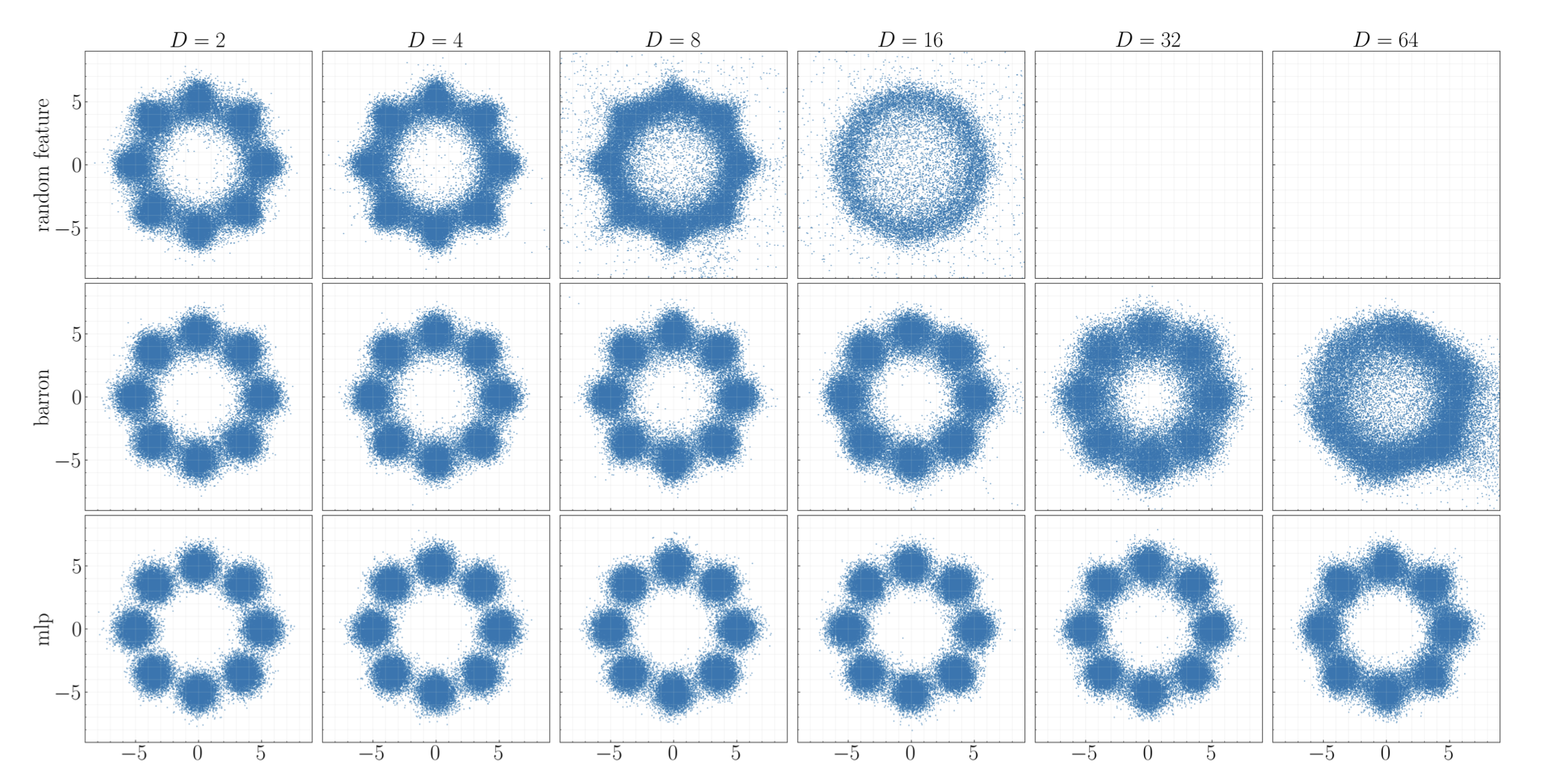
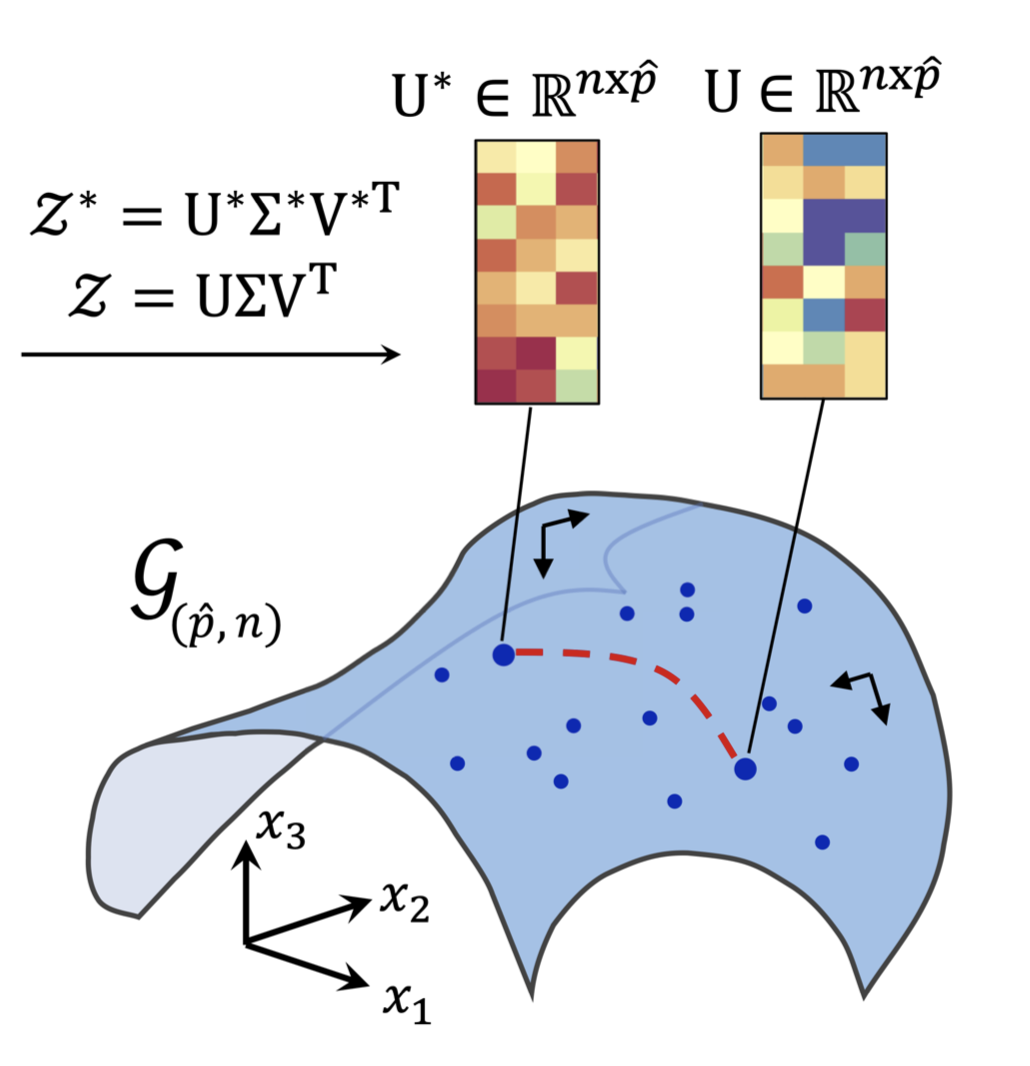
- Methods for accelerated generative modeling over geometric data, combining flow maps with Riemannian geometry for proteins and molecules
Current topics of interest include 3D shape generation, molecular sampling, scientific inverse problems, and rare event estimation in complex dynamical systems.
Beyond our core methodological work, we are early adopters of AI tools and are excited about a future of scientific research deeply integrated with AI. We investigate new ways to leverage language models and agentic automation to dramatically accelerate the pace of research within our group and beyond.
PhD Students
Jerry Huang
Computer Science Department
Stephen Huan
Computer Science Department
co-advised with Andrej Risteski
Masters Students
Kartik Nair
Machine Learning Department
Sheel Shah
Machine Learning Department
Undergraduate Students
Ishin Shah
Department of Mathematical Sciences
co-advised with Max Simchowitz
Publications
Code
We are strong advocates for reproducible science and open-source software. We believe that they have been key drivers of the recent advances in AI research, and we make all our code publicly available to continue this tradition.
-
jax-interpolants
A JAX implementation of the stochastic interpolant framework for generative modeling
-
jax-edm2
A JAX implementation of NVIDIA's EDM2 U-Net architecture
-
flow-maps (with Michael Albergo and Eric Vanden Eijnden)
A JAX implementation of the self-distillation framework for learning flow maps and consistency models
-
model-free probability flows (with Eric Vanden Eijnden)
Code for model-free learning of probability flows in nonequilibrium dynamics
-
structured diffusion (with Arthur Jacot and Stephen Tu)
Experiments verifying provable guarantees for when shallow diffusion networks can learn hidden structure
-
flow map matching (with Michael Albergo and Eric Vanden Eijnden)
A JAX implementation of the flow map matching framework for learning flow maps and consistency models
-
forecasting with interpolants (with Mengjian Hua, Yifan Chen, Mark Goldstein, Eric Vanden Eijnden, and Michael Albergo -- mostly written by the first three authors!)
An implementation of probabilistic forecasting with stochastic interpolants
-
active probability flows (with Eric Vanden Eijnden)
Code for learning probability flows and entropy production rates in active matter
-
stochastic interpolants (with Michael Albergo and Eric Vanden Eijnden)
An implementation of the unifying stochastic interpolant framework for flows and diffusions in generative modeling
-
score-based transport modeling (with Eric Vanden Eijnden)
Code for constructing a probability flow solution of the Fokker-Planck equation
-
spin glass evolutionary dynamics (with Yipei Guo, Chris Rycroft, and Ariel Amir)
High-resolution C++ simulation code to model Lenski's Long-Term Evolution Experiment, taking into account microscopic epistasis and clonal interference in microbial evolution
-
shear transformation zone++ (with Chris Rycroft)
Three-dimensional MPI-based C++ code for parallel simulation of quasi-static deformation in elastoplastic solids
-
real-space Hartree-Fock (with Amir Natan, note: only the Hartree-Fock implementation)
Parallelized Fortran code to efficiently compute the Hartree-Fock exchange via the use of projection operators onto occupied and virtual states
Teaching
I teach mathematically- and computationally-oriented courses in applied mathematics and machine learning. Below is a list of past, present, and future offerings.
Carnegie Mellon, Spring 2025: Methods of Optimization
Carnegie Mellon, Fall 2024: Introduction to PDEs: A Computational Approach
NYU Courant, Spring 2024: Honors Numerical Analysis
NYU Courant, Fall 2023: Linear and Nonlinear Optimization
NYU Courant, Spring 2023: Linear and Nonlinear Optimization
NYU Courant, Fall 2022: Numerical Analysis
NYU Courant, Spring 2022: Linear and Nonlinear Optimization
NYU Courant, Fall 2021: Numerical Analysis
Northwestern, Full Year 2014: Introduction to Computer Programming for Integrated Science